User:David Canner/Sandbox hmgr
From Proteopedia
|
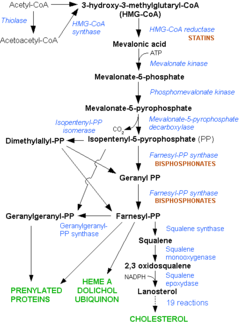
HMG-CoA Reductase (or 3-hydroxy-3-methyl-glutaryl-CoA reductasese or HMGR) is the rate-controlling enzyme of the metabolic pathway responsible for cholesterol and other isoprenoid biosynthesis, the mevalonate pathway. HMGR is a polytopic, transmembrane protein, containing 8 domains, that is anchored in the membrane of the endoplasmic reticulum.[1] It is the major target of the Statins, a cholesterol lowering drug class and the best selling pharmaceutical drugs in the world.
Biological RoleHMGR is among the most highly regulated enzymes in the human body. It catalyzes the formation of mevalonic acid, the committed step in the biosynthesis of sterols, notably cholesterol and isoprenoids. This reaction can be seen below where HMG-CoA is reduced by NADPH. Despite the poor reputation cholesterol has in the media, it is a critical component of cellular membranes as it is required to establish proper membrane permeability and fluidity. The mevalonate pathway is also responsible for synthesis of the oxygen transporting heme found in red blood cells. [2]
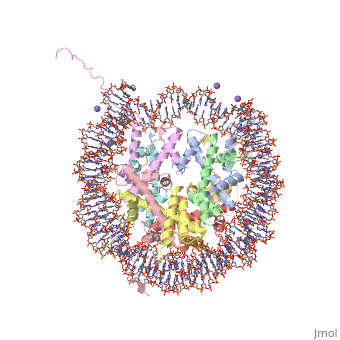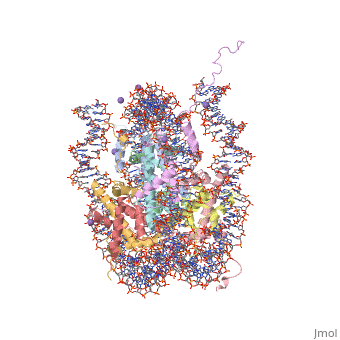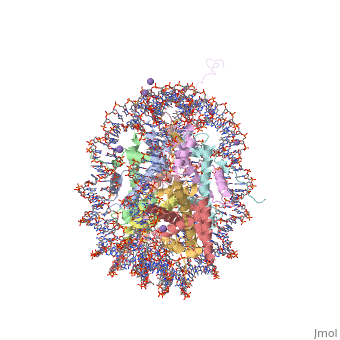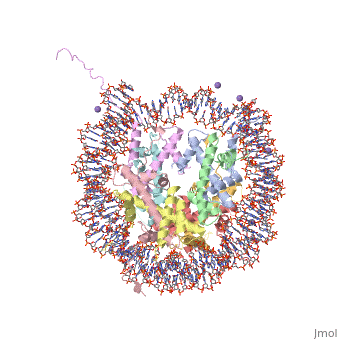
StructureGeneral StructureThere are two distinct classes of HMGRs, class I, which is only found in eukaryotes and are membrane bound and class II, which is found in prokaryotes and are soluble. [3] HMGR contains 8 transmembrane domains, which have yet to be successfully crystallized, that anchor the protein to the membrane of the endoplasmic reticulum. [1] The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other. [1] Within the tetramer, the monomers are arranged into , each of which contains which are formed by residues form both monomers. Each monomer contains , the , the , and the . The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of ferredoxin. The S and L domains are connected by a which is essential for the HMG-binding site. [1] Salt bridges between residues R641 and E782 as well as between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface. [1]
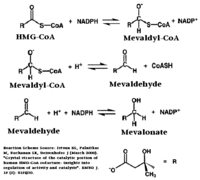
Substrate Binding & Catalytic MechanismThe HMG-CoA and NADPH molecules make numerous contacts with the L and S domains in forming the four active sites. The CoA is located in a , with the pantothenic acid moiety extending into the interior of the protein. over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with playing a critical role in binding. [1] The HMG binding pocket is the site of catalysis in HMGR. is a critical structural element of this binding site. Residues and are positioned in the active site as is . It is this K691 that presumably stabilizes the negatively charged oxygen on the first mevaldyl-CoA intermediate. [1] The mevaldyl CoA intermediate is subsequently converted to Mavaldehyde with added stabilization from . It is then believed that the close proximity of increases the pKA of E559, allowing it to be a proton donor for the reduction of mevaldehyde into mevalonate. [1]
Regulation of HMG-CoA ReductaseAs stated before, HMGR is among the most highly regulated enzymes in the human body. [2] Regulation of HMG-CoA reductase occurs at 4 different levels of feedback regulation. First by regulation of transcription of the reductase gene, which is activated by sterol regulatory element binding protein, a protein that binds to the promoter of the HMGR gene when cholesterol levels fall. The second level of regulation is at the translation of the HMGR mRNA, which is inhibited by Farnesol, a derivative of the mevalonate pathway. [2] The third level of HMGR regulation involves the degradation of intact reductase. As the level of sterols increases, HMGR reductase becomes more susceptible to ER-associated degradation. Helices 2-6 of the HMGR transmembrane domain, called the sterol-sensing domain, sense the increased levels of sterols. Degradation of HMGR is initiated when a membrane-bound cysteine protease cleaves within the transmembrane region of HMGR. Additionally, as sterol levels increase, the helices can slightly rearrange, exposing Lysine 248 which can be ubiquitinated and subsequently trigger proteolytic degradation. [1] [4] A final level of HMGR regulation is achieved by inhibition via . HMGR is phosphorylated and inactivated by an AMP-activated protein kinase, when the energy charge of the cell is low and AMP concentrations are high. Since Serine 872 is in the vicinity of the pohsphate of NADP, phosphorylation likely results in a decreased affinity for NADPH, halting the formation of Mevaldyl-CoA from HMG-CoA.
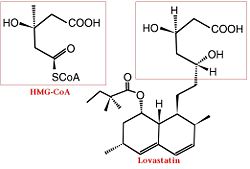
[5] Medical ImplicationsElevated cholesterol levels have been identified as a major risk factor for coronary artery disease, the narrowing of arteries of the heart, which affected over 13 million people in the United States alone. It is a major cause of disability and death, killing over 500 thousand people in the USA in 2001. [6] The statins are HMG-CoA reductase inhibitors. Discovered by Akira Endo in 1971 [7], stains are similar in structure to HMG-CoA and act by competitively inhibiting HMGR. Since HMGR is the first committed enzyme in the cascade that eventually produces cholesterol, use of statins can dramatically reduce blood cholesterol levels. As a drug class, statins generated over $20 billion dollars in sales in 2009 with Pfizer’s lipitor the best selling drug in the world. [8][9] A number of crystal structures of HMGR with bound statins have been solved which elucidate how the statin molecule is bound by HMGR. Statins in general occupy the active site of HMGR, preventing HMG-CoA from being bound by the enzyme. The structures of HMGR with bound , , , , , and highlight the important residues involved in inhibitor binding. The cis loop forms a number of polar interactions with the inhibitor, particularly residues Ser 684, Asp 690, Lys 691, Lys 692, and hydrogen bond interactions between Glu 559 and Asp 767 with the O5-hydroxyl of the statins. Van der Waals interactions between Leu 562, Val 683, Leu 853, Ala 856, and Leu 857 of HMGR and hydrophobic ring structures of the statins. [10]These interactions serve to allow the statins to be bound by HMGR with a Ki of between .1-2.3nM while the Michaelis constant for HMG-CoA is 4uM. [11]
Additional 3D Structures of HMG-CoA Reductase3cct, 3ccw, 3ccz, 3cd0, 3cd5, 3cd7, 3cda, 3cdb – HMG-CoA Reductase Catalytic Domain Additional Resources
References
|