User:Madelyn Kasprzak/Sandbox 6
From Proteopedia
Contents |
TET Enzymes
TET enzymes are a family of dioxygenases that are involved in the process of oxidizing methylated cytosine. Members of this family include ten-eleven translocation methylcytosine dioxygenase 1 (), methylcytosine dioxygenase , and methylcytosine dioxygenase . The gene for the first of these proteins, TET1, was identified when it was determined to be fused to the Mixed Lineage Leukemia (MLL) gene as a result of a translocation event that occurred between chromosomes ten and eleven (hence the name). [1]
Structure
The TET enzymes have a cysteine-rich region closely followed by a double stranded beta-helix (DSBH) domain near their C-terminus.[2] The DSBH domain contains three and an .[2] For all α-ketoglutarate oxygenases, including TET enzymes, the DSBH domain and the preceding cysteine-rich region perform the main catalytic activity for these enzymes.
In addition, TET1 has a CXXC-type zinc finger domain near the N-terminus. However, the TET1 CXXC domain lacks the conserved lysine-phenylalanine-glycine-glycine (KFGG) motif commonly seen within the CXXC domains of other DNA binding proteins, such as DNA methyltransferase-1 (DNMT1). A study conducted by Frauer et al. in 2011 showed that the isolated CXXC domain of TET1 has no DNA binding activity, which agrees with the evidence suggesting that the KFGG motif increases affinity for unmethylated DNA.[3] Frauer et al. also speculated that the CXXC domain of TET1 may be involved with protein-protein interactions instead of DNA binding.[3]
|
Function
Common Function
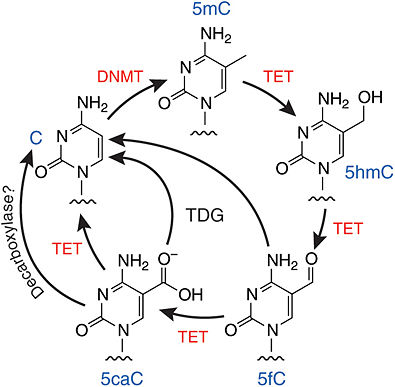
All three TET enzymes and their isoforms are involved in the biochemical pathway that converts 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC). They also regulate the further conversions of 5hmC to 5-formylcytosine (5fC) and then 5fC to 5-carboxylcytosine (5caC).[4] Although experimental data shows that TET3 does so to a lesser extent than TET1 and TET2.[4]
While the oxidation performed by TET enzymes was originally thought to be a source of DNA damage, new research has implied that this catalytic activity may actually be the initial steps of a process of DNA demethylation. This hypothesized DNA demethylation pathway starts with the conversion of 5mC to 5caC after several rounds of oxidation by TET enzymes. The next step is the removal of the modified cytosine base by thymine DNA glycosylase (TDG) which leaves an abasic site on the DNA. The last step is then the process of base excision repair in which a new unmodified cytosine is regenerated at the site, thus completing the process of DNA demethylation.[4][5]
Specific Functions
Experimental data shows that the TET genes have different expression patterns and at different levels, which indicates that each of the TET enzymes do not fully overlap in their functionality.
TET1 is usually expressed in fetal heart, lung, and brain tissue and in adult skeletal muscle, thymus, and ovary. It is not generally expressed in adult heart, lung, and brain tissue. Moreover, studies have shown that TET1 expression in adult brain tissue is correlated with brain cancer. This occurs through TET1’s indirect activation of brain cancer-related genes such as EGFR, AKT3, CDK6, CCND2, and BRAF through creation of 5hmC which recruits the CHTOP-methylosome complex that activates these genes.[7]
TET2 is broadly expressed, but it is especially highly expressed in hematopoietic cells, which develop into blood cells. Regarding this, it is suggested that TET2 plays a role in hematopoiesis due to the presence of TET2 mutations in many myelodysplastic syndromes.[8][9]
TET3 is highly expressed in zygotes and is involved with epigenetic chromatin reprogramming in the zygote after fertilization. Specifically, it plays a role in DNA demethylation of the paternal pronucleus before implantation.[5]
Disease Relation
TET1 Isoform
Point mutations within the TET1 isoform can lead to a loss of enzyme function which causes a lack of DNA demethylation; two occur at the 1672 and 1674 amino acid residues, the first being a H1672Y mutation and the second being a D1674A mutation. A severe mutation in the 1608-1609 codons can lead to TET1 becoming an oncogene in acute leukemias. This mutation fuses TET1 and KMT2A/MLL1 to form an oncogene.[10][11]
TET2 Isoform
TET2 has been identified as a tumor-suppressor gene that is linked to various myeloid cancers. Acquired mutations are predicted to truncate the protein, resulting in alterations of the function of the TET2 protein. TET2 defects have been identified in the CD34+ cells of patients with myelodysplastic syndrome, which include hematopoietic stem cells and hematopoietic progenitors. This suggests that TET2 mutations or defects, paired with mutations in other genes, such as Janus kinase 2 (JAK2) V617F gene or myeloproliferative leukemia virus oncogene (MPL) W515L/K can contribute to the progression of myeloid cancers. Due to the involvement of TET2 in hematopoietic stem cell development, mutations in this gene may also be associated with the amplification of the mutation in the early stages of hematopoietic differentiation.[8]
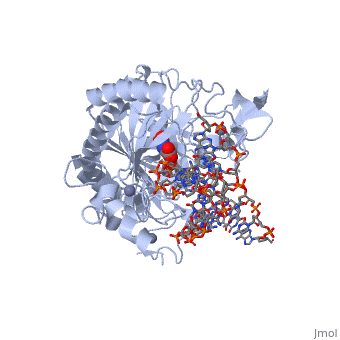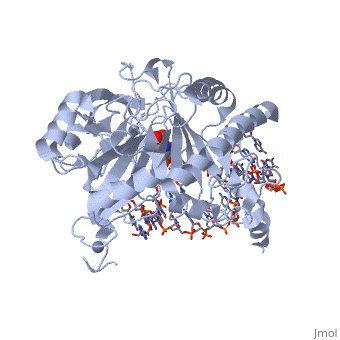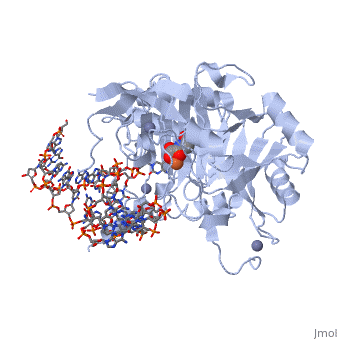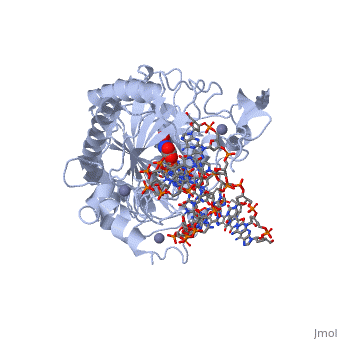
Structural highlights
The following highlights are of the Human TET2 enzyme in complex with DNA.
are colored red.
are colored green.
Both are displayed in this scene.
Here the are colored light blue, but since they are so small they may be slightly difficult to see.
Here some of the major residues of the are highlighted. The light green residues are 2-oxoglutarate binding residues, the orange residue binds the substrate, and the blue residue binds the catalytic iron ion. For the rest of the protein, only the backbone is shown in light gray.
References
- ↑ Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003 Mar;17(3):637-41. PMID:12646957 doi:http://dx.doi.org/10.1038/sj.leu.2402834
- ↑ 2.0 2.1 Kinney SR, Pradhan S. Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer. Adv Exp Med Biol. 2013;754:57-79. doi: 10.1007/978-1-4419-9967-2_3. PMID:22956496 doi:http://dx.doi.org/10.1007/978-1-4419-9967-2_3
- ↑ 3.0 3.1 Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, Wang M, Qin W, Soding J, Spada F, Leonhardt H. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011 Feb 2;6(2):e16627. doi: 10.1371/journal.pone.0016627. PMID:21311766 doi:http://dx.doi.org/10.1371/journal.pone.0016627
- ↑ 4.0 4.1 4.2 He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011 Sep 2;333(6047):1303-7. doi: 10.1126/science.1210944. Epub 2011 Aug, 4. PMID:21817016 doi:http://dx.doi.org/10.1126/science.1210944
- ↑ 5.0 5.1 Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013 Oct 24;502(7472):472-9. doi: 10.1038/nature12750. PMID:24153300 doi:http://dx.doi.org/10.1038/nature12750
- ↑ Huang Y, Rao A. New functions for DNA modifications by TET-JBP. Nat Struct Mol Biol. 2012 Nov;19(11):1061-4. doi: 10.1038/nsmb.2437. PMID:23132381 doi:http://dx.doi.org/10.1038/nsmb.2437
- ↑ Takai H, Masuda K, Sato T, Sakaguchi Y, Suzuki T, Suzuki T, Koyama-Nasu R, Nasu-Nishimura Y, Katou Y, Ogawa H, Morishita Y, Kozuka-Hata H, Oyama M, Todo T, Ino Y, Mukasa A, Saito N, Toyoshima C, Shirahige K, Akiyama T. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014 Oct 9;9(1):48-60. doi: 10.1016/j.celrep.2014.08.071. Epub 2014 Oct, 2. PMID:25284789 doi:http://dx.doi.org/10.1016/j.celrep.2014.08.071
- ↑ 8.0 8.1 Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009 May 28;360(22):2289-301. doi: 10.1056/NEJMoa0810069. PMID:19474426 doi:http://dx.doi.org/10.1056/NEJMoa0810069
- ↑ Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009 Jul;41(7):838-42. doi: 10.1038/ng.391. Epub 2009 May 31. PMID:19483684 doi:http://dx.doi.org/10.1038/ng.391
- ↑ Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002 Jul 15;62(14):4075-80. PMID:12124344
- ↑ Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003 Mar;17(3):637-41. PMID:12646957 doi:http://dx.doi.org/10.1038/sj.leu.2402834