T. brucei Phosphodiesterase B1
From Proteopedia
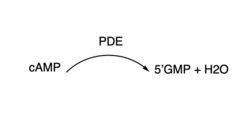
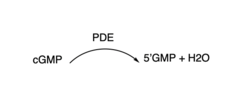
3′,5′-cyclic nucleotide phosphodiesterases (PDEs) are enzymes that hydrolyse 3',5'-phosphodiester bonds in two of the most important cell signalling molecules, cyclic-adenosine monophosphates (cAMPs) and cyclic-guanosine monophosphates (cGMPs). PDEs are essential for the inactivation of cAMP and or cGMP by controlling the rate at which they are degraded to regulate their intracellular concentrations and maintain cellular homeostasis [1]. PDEs directly modulate these messenger molecules by degrading cAMP or cGMP when concentrations are elevated beyond the cellular threshold [2]. These enzymes are involved in a wide array of cellular and physiological processes such as metabolism, some immune responses, reproduction, inflammation, regulation of photoreceptors, quorum sensing in prokaryotes, enabling parasite transition through life cycles stages and the list goes on [3].
Contents |
Function
is categorized as a class I enzyme, a class which currently is comprised of all known mammalian PDE enzymes. PDEB1 is one of a few cyclic nucleotide phosphodiesterases (others include, TbPDE2C, TbrPDEB2) that have been validated as promising drug targets for the treatment of Human African Trypanosomiasis (HAT) also known as sleeping sickness, a life-threatening infectious disease which is caused by Trypanosoma brucei parasites [4]. Trypanosomes express four different PDEs (PDE A-D) each capable of exiting in different isoforms with catalytic domains that are highly similar to the catalytic domain of human PDE [5]. In T. brucei, PDEB1 is a cAMP-specific hydrolase that catalyze the hydrolysis of cAMP to AMP within the catalytic domain of the enzyme. TbPDEB1 enzyme has been reported to play a major role in facilitating Trypanosoma motility in host cells and subsequent progression through the parasite life cycle [6]. On the other hand, TbPDE2C has been implicated in regulating the proliferation of T. brucei parasites during the bloodstream stage of infection after a study observed that cAMP accumulation (due to PDE inhibition) is toxic to bloodstream Trypanosomes [7].
PDEs can either be cAMP, cGMP or dual-to cAMP and cGMP- specific, and the substrate specificity for the enzyme is a very important consideration in determining enzyme-inhibitor selectivity and specificity. That is, some PDE inhibitors may have higher binding affinity to cAMP-PDEs than to cGMP-PDEs, or similar binding interactions to dual substrate-specific enzymes [8].
Relevance
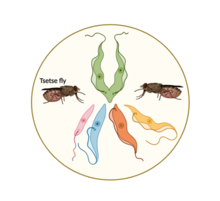
Trypanosomiasis, also known as HAT, or sleeping sickness, is a neglected tropical disease that is fatal if left untreated and is one of the major contributors to human and livestock mortality in developing countries. HAT is transmitted by blood-sucking female Tsetse flies infected with trypanosomes. Trypanosomes undergo some of their complex life cycle stages in the tsetse fly vector until they reach the infective stage where they are then transferred to the host, human or livestock animals. Trypanosomiasis takes two forms depending on the subspecies of infection agent, ie. trypanosomes. One form leads to chronic infection and or sleeping, it is caused by T. brucei gambiense and it is implicated in about 95% of trypanosomiases cases. The other rarer form leads to a a more severe clinical presentation characterized by an acute, rapid infection which invades the central nervous system (CNS). While trypanosomiasis is primarily endemic to some countries in west and central Africa, it also affects about 72% of European and American tourists who visit safari reserves and game parks in endemic regions [9]. HAT can be transmitted congenitally, i.e. from mother to child, through mechanical transmission (insect bite), blood transfusions or contaminated needles, and through sexual intercourse. During the first stage of infection, also called the haemo-lymphatic or simply bloodstream stage, trypanosomes multiply in subcutaneous tissues, lymph and blood which leads to symptoms like headaches, fever, headaches, itching, swollen lymph nodes, and joint pains. The second stage of infection is called the meningo-encephalic stage, this is when the parasites successfully cross the blood-brain barrier and infect the central nervous system. Symptoms of trypanosomiasis are more apparent in this stage and may include confusion, behavioural changes, loss of coordination and disturbance of sleep-cycle hence the disease name. Some rare cases of healthy carriers of the disease have been reported, however, without proper treatment, sleeping sickness is largely fatal [10].
The current treatment options for trypanosomiasis are outdated - such as melarsoprol which was first introduced in 1949- and sub-optimal typically showing high toxicity, particularly in late-stage infections [11]. There is a clear need for safer, more efficacious drugs to treat sleeping sickness which is why creative studies are being performed to validate new drug targets for trypanosomiasis drug development. PDEB1 makes an interesting drug target for HAT drug discovery due to its essential functions that contribute to parasite fitness and survival. Other PDEs in trypanosomes such as PDEB2, PDE2A, PDE2C, are also under evaluation as potential drug targets due to their specific function in regulating different stages of the parasite life cycle.
Structural highlights
|
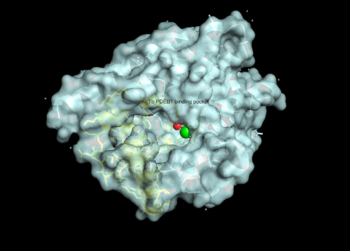
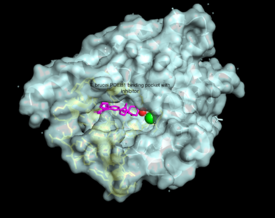
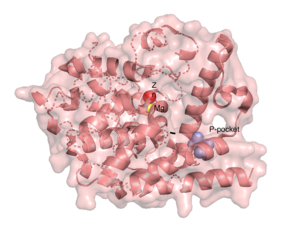
There are several resolved crystal structures for TbPDEB1 in both apo and holo forms which facilitate virtual screenings of libraries of compounds for inhibitory activity against TbPDEB1. The active site for TbPDEB1 contains Magnesium (Mg) and Zinc (Zn) ions with each metal ion forming an octahedral geometry. The metal ion coordinates side chains of two histidine residues and two aspartic residues and ion coordinates D710 and five water molecules.[12]. Only the catalytic domain of the TbPDEB1 enzyme performs hydrolysis of cAMP to AMP [13]. The solved crystal structure of TbPDEB1 enabled the visualization of enzyme ligand and metal binding pocket, which creates a platform for virtual drug screening against the target. That is, compounds can be rapidly selected based on their ability to fit within the binding pocket, and improvements can be made to compounds to support tighter binding. In addition, there is a (residues HNFYH, position 669-673) which facilitates the binding of substrate, cAMP, and the subsequent increase in catalytic activity.
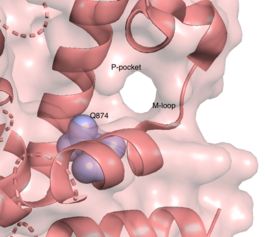
The resolved crystal structure for the catalytic domain of TbPDEB1 (PDB identifier: 4I15) confirmed the existence of a T. brucei specific , which has since proven to be an exciting direction for improving drug selectivity for T. brucei phosphodiesterase. Following the identification of the parasite specific P-pocket just adjacent to the enzyme binding pocket, new TbPDEB1 inhibitor analogues were created to extend into the parasite enzyme p-pocket. This resulted in improved selectivity for T. brucei PDEB1 over human PDE4, another cAMP specific phosphodiesterase.
Alpha fold structure prediction
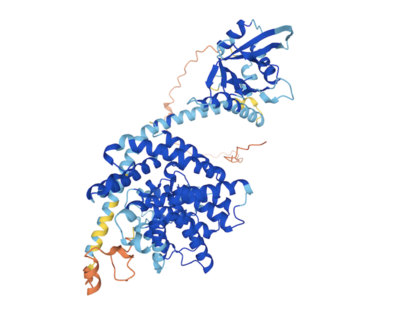
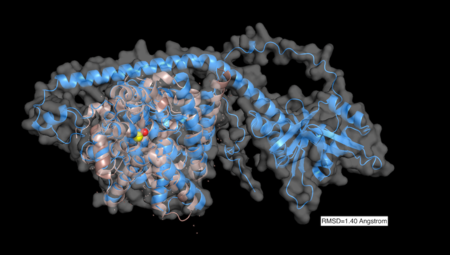
The prediction structure overall showed high confidence especially in the enzyme catalytic domain which yielded structure prediction with very high confidence (pLDDT > 90) [14] [15] . When AlphaFold predicted structure for TbPDEB was aligned with unliganded crystal structure for , overlap was observed between the catalytic domain of the crystal structure and the catalytic domain of the predicted structure (see 2D images below illustrating AlphaFold structure prediction confidence with deep blue designated as very high confidence and Pymol [16] alignment result).
Other liganded TbPDEB crystal structures [17]
4I15: Crystal structure of unliganded T. brucei PDEB1
5L8C: Structure based identification of selective inhibitors of TbPDEB1
4PHL: Structure based determination of selective inhibitors of TbPDEB1
5G57: Structure based determination of selective inhibitors of TbPDEB1
5G5V: Structure based determination of selective inhibitors of TbPDEB1
Protein evolution conservation profile
Citations
- ↑ Baker DA. Cyclic nucleotide signalling in malaria parasites. Cell Microbiol. 2011 Mar;13(3):331-9. doi: 10.1111/j.1462-5822.2010.01561.x. Epub, 2010 Dec 28. PMID:21176056 doi:http://dx.doi.org/10.1111/j.1462-5822.2010.01561.x
- ↑ Blaazer AR, Singh AK, de Heuvel E, Edink E, Orrling KM, Veerman JJN, van den Bergh T, Jansen C, Balasubramaniam E, Mooij WJ, Custers H, Sijm M, Tagoe DNA, Kalejaiye TD, Munday JC, Tenor H, Matheeussen A, Wijtmans M, Siderius M, de Graaf C, Maes L, de Koning HP, Bailey DS, Sterk GJ, de Esch IJP, Brown DG, Leurs R. Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity. J Med Chem. 2018 May 1. doi: 10.1021/acs.jmedchem.7b01670. PMID:29672041 doi:http://dx.doi.org/10.1021/acs.jmedchem.7b01670
- ↑ Zoraghi R, Seebeck T. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc Natl Acad Sci U S A. 2002 Apr 2;99(7):4343-8. PMID:11930001 doi:http://dx.doi.org/10.1073/pnas.062716599
- ↑ Jansen C, Wang H, Kooistra AJ, de Graaf C, Orrling KM, Tenor H, Seebeck T, Bailey D, de Esch IJ, Ke H, Leurs R. Discovery of Novel Trypanosoma brucei Phosphodiesterase B1 Inhibitors by Virtual Screening against the Unliganded TbrPDEB1 Crystal Structure. J Med Chem. 2013 Mar 1. PMID:23409953 doi:http://dx.doi.org/10.1021/jm3017877
- ↑ Tagoe DN, Kalejaiye TD, de Koning HP. The ever unfolding story of cAMP signaling in trypanosomatids: vive la difference! Front Pharmacol. 2015 Sep 7;6:185. doi: 10.3389/fphar.2015.00185. eCollection, 2015. PMID:26441645 doi:http://dx.doi.org/10.3389/fphar.2015.00185
- ↑ Shaw S, DeMarco SF, Rehmann R, Wenzler T, Florini F, Roditi I, Hill KL. Flagellar cAMP signaling controls trypanosome progression through host tissues. Nat Commun. 2019 Feb 18;10(1):803. doi: 10.1038/s41467-019-08696-y. PMID:30778051 doi:http://dx.doi.org/10.1038/s41467-019-08696-y
- ↑ Zoraghi R, Seebeck T. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc Natl Acad Sci U S A. 2002 Apr 2;99(7):4343-8. PMID:11930001 doi:http://dx.doi.org/10.1073/pnas.062716599
- ↑ Baker DA. Cyclic nucleotide signalling in malaria parasites. Cell Microbiol. 2011 Mar;13(3):331-9. doi: 10.1111/j.1462-5822.2010.01561.x. Epub, 2010 Dec 28. PMID:21176056 doi:http://dx.doi.org/10.1111/j.1462-5822.2010.01561.x
- ↑ Kennedy PGE, Rodgers J. Clinical and Neuropathogenetic Aspects of Human African Trypanosomiasis. Front Immunol. 2019 Jan 25;10:39. doi: 10.3389/fimmu.2019.00039. eCollection, 2019. PMID:30740102 doi:http://dx.doi.org/10.3389/fimmu.2019.00039
- ↑ Ponte-Sucre A. An Overview of Trypanosoma brucei Infections: An Intense Host-Parasite Interaction. Front Microbiol. 2016 Dec 26;7:2126. doi: 10.3389/fmicb.2016.02126. eCollection, 2016. PMID:28082973 doi:http://dx.doi.org/10.3389/fmicb.2016.02126
- ↑ Kennedy PG. Human African trypanosomiasis of the CNS: current issues and challenges. J Clin Invest. 2004 Feb;113(4):496-504. doi: 10.1172/JCI21052. PMID:14966556 doi:http://dx.doi.org/10.1172/JCI21052
- ↑ Jansen C, Wang H, Kooistra AJ, de Graaf C, Orrling KM, Tenor H, Seebeck T, Bailey D, de Esch IJ, Ke H, Leurs R. Discovery of Novel Trypanosoma brucei Phosphodiesterase B1 Inhibitors by Virtual Screening against the Unliganded TbrPDEB1 Crystal Structure. J Med Chem. 2013 Mar 1. PMID:23409953 doi:http://dx.doi.org/10.1021/jm3017877
- ↑ Kunz S, Luginbuehl E, Seebeck T. Gene conversion transfers the GAF-A domain of phosphodiesterase TbrPDEB1 to one allele of TbrPDEB2 of Trypanosoma brucei. PLoS Negl Trop Dis. 2009 Jun 9;3(6):e455. doi: 10.1371/journal.pntd.0000455. PMID:19513125 doi:http://dx.doi.org/10.1371/journal.pntd.0000455
- ↑ Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Jul 15. pii: 10.1038/s41586-021-03819-2. doi:, 10.1038/s41586-021-03819-2. PMID:34265844 doi:http://dx.doi.org/10.1038/s41586-021-03819-2
- ↑ Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Zidek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022 Jan 7;50(D1):D439-D444. doi: 10.1093/nar/gkab1061. PMID:34791371 doi:http://dx.doi.org/10.1093/nar/gkab1061
- ↑ Schrödinger LLC & DeLano, W. (2020). PyMOL. Retrieved from http://www.pymol.org/pymol
- ↑ Blaazer AR, Singh AK, de Heuvel E, Edink E, Orrling KM, Veerman JJN, van den Bergh T, Jansen C, Balasubramaniam E, Mooij WJ, Custers H, Sijm M, Tagoe DNA, Kalejaiye TD, Munday JC, Tenor H, Matheeussen A, Wijtmans M, Siderius M, de Graaf C, Maes L, de Koning HP, Bailey DS, Sterk GJ, de Esch IJP, Brown DG, Leurs R. Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity. J Med Chem. 2018 May 1. doi: 10.1021/acs.jmedchem.7b01670. PMID:29672041 doi:http://dx.doi.org/10.1021/acs.jmedchem.7b01670