Atomic coordinate file
From Proteopedia
Contents |
Definition
Atomic coordinate files are the data files that specify three-dimensional (3D) molecular structures. At a minimum, they must specify the positions of each atom in space, typically with X, Y and Z Cartesian coordinates, and the chemical element each atom represents.
Data Formats
Atomic coordinate files use many possible data formats. The XYZ format (file type .xyz) specifies only the coordinates and chemical element for each atom, and is useful for small molecules. This format is not adequate for macromolecules because additional information is needed for their atoms.
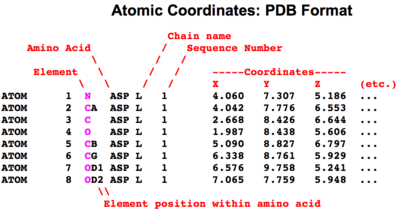
Macromolecular atomic coordinate files need to specify quite a bit of information in addition to the position of each atom in space and its chemical element. Each atom either belongs to a Standard Residue or not. If not, it is designated a hetero atom. The position of each atom within a standard residue is specified, e.g. carbon atoms in amino acids can be the carboxy carbon (C), the alpha carbon (CA), the beta carbon (CB), and so forth. Nitrogen atoms can be in the main chain (N), or on the sidechain, e.g. in the terminal zeta position in lysine (NZ). In addition to the name of the residue to which an atom belongs are provided the name of the chain where the residue is found, and its sequence number position. In addition to the X, Y, and Z coordinates are given an occupancy value, and an isotropic B value or temperature value''.
PDB Data Format
The most popular macromolecular data format among crystallographers is the one developed and used by the early (1970's) Protein Data Bank, called the Protein Data Bank Format, PDB Format, or legacy PDB format. Data files in this format are called PDB Files (file type .pdb). Although this format has serious limitations, it remains popular partly because the data files are in plain text, and are relatively easy to read by humans.
PDB format cannot accommodate >99,999 atoms/model, or >62 chains (see Jmol/Visualizing large molecules). In August, 2021, the PDB format accommodates >99% of X-ray crystallography entries, but only about 86% of cryo-EM entries[1]. The remainder are available in mmCIF format (see below). 88% of entries were determined by X-ray, and 4.5% by cryo-EM. For the entire database as a whole, 98.8% of entries are available in PDB format (August, 2021).
|
Simplified Diagram of ATOM Records in the PDB Format. Not shown (under etc.) are the occupancy and temperature value. ENLARGE. |
To view a PDB file from a PDB code-titled page in Proteopedia, click on the OCA link beneath the molecule. At OCA, scroll down to the Data Retrieval section, and click on complete with coordinates in the first line there.
To view the text of a PDB file at the RCSB PDB, go to the page for the PDB identification code of interest, then at the upper right, click Display Files, and under that heading, PDB File.
- Simple Diagram of ATOM Records in the PDB Format (see also HETATM)
- Protein Data Bank PDB Format Description
Retirement of PDB Format
In February, 2019, the wwPDB announced that new depositions must be in the mmCIF format beginning July 1, 2019[2]. The PDB sometimes refers to the mmCIF format as "PDBx", which should not be confused with the original legacy PDB format.
In December, 2023, the wwPDB announced that all 3-character ligand ID codes had been exhausted [3]. Thereafter, new entries with novel ligands will be available only in mmCIF format, since the legacy PDB format cannot accommodate the new 5-character ligand IDs. Examples that use 5-character ligand IDs: 8rox has A1H17; 8vkz has A1ACE.
In 2024, the wwPDB estimates that all 4-character PDB ID codes will be consumed by 2029[4]. Thereafter, new entries will be available only in mmCIF format using 12-character ID codes.
mmCIF Data Format
In response to the inadequacies of the PDB data format, the International Union of Crystallographers and the World Wide Protein Data Bank have adopted the macromolecular crystallographic information format (mmCIF) as their primary data format for macromolecules. mmCIF is also sometimes referred to as PDBx (not to be confused with the PDB format). While the mmCIF/PDBx format has considerable merit from the perspective of computer scientists, it is unpopular with crystallographers, who prefer to work in the PDB data format. Therefore, the PDB has maintained the entire database in both formats. However, new depositions must be in the mmCIF format beginning July 1, 2019, and it is anticipated that the PDB format will be phased out, of necessity, around 2026[2][5].
Models Available Only in mmCIF Format
In April, 2024, 2.3% of the entries in the wwPDB are available only in mmCIF format.
Models with >99,999 atoms, or >62 chains, do not fit in the PDB format (see Jmol/Visualizing large molecules). Such models are available only in mmCIF format, and not in the PDB format. However, in 2024, such models are available in subsets in PDB format. For example, at 5LEG, look for "PDB format-like files" in the Download Files menu.
Models containing ligands with 5-character ID codes (see above) also do not fit in PDB format, and are available only in mmCIF format.
ASN.1 Data Format
The US National Center for Biotechnology Information (NCBI) maintains a macromolecular structure database (derived from the Protein Data Bank) that is integrated with their Entrez cross-database search system, and their other databases of sequences, medical literature, inheritance, taxonomy, etc. They have chosen to maintain their atomic coordinate files in the Abstract Syntax Notation One (ASN.1) data format.
Bonds: Connectivity
Typically, atomic coordinate files do not specify covalent bonds between atoms. Molecular modeling or visualization software determines the positions of covalent bonds using simple rules. Typically, any two non-hydrogen atoms within 1.9 Ångstroms of each other are deemed to be covalently bonded. (The distance for a bond involving a hydrogen atom is less.) The PDB data format requires that covalent bonds be specified between atoms that are not members of Standard Residues in protein or nucleic acid chains. These are specified in CONECT records.
See Also
- Protein Data Bank
- PDB identification code
- Standard Residues
- Non-Standard Residues
- Hetero atoms
- Ligand
- Temperature value
Notes and References
- ↑ The advanced search at RCSB.org has a field Deposition, Compatible with PDB format.
- ↑ 2.0 2.1 Mandatory PDBx/mmCIF format files submission for MX depositions: posted on the PDB email list Feb 20, 2019 by Jasmine Young, Biocuration Team Lead, RCSB PDB. The wwPDb website also posted this document.
- ↑ PDB Entries with Novel Ligands Now Distributed Only in PDBx/mmCIF and PDBML File Formats, wwPDB News, December 12, 2023.
- ↑ Resources for Supporting the Extended PDB ID Format (pdb_00001abc), Spring 2024 Issue of the RCSB PDB Newsletter.
- ↑ Adams PD, Afonine PV, Baskaran K, Berman HM, Berrisford J, Bricogne G, Brown DG, Burley SK, Chen M, Feng Z, Flensburg C, Gutmanas A, Hoch JC, Ikegawa Y, Kengaku Y, Krissinel E, Kurisu G, Liang Y, Liebschner D, Mak L, Markley JL, Moriarty NW, Murshudov GN, Noble M, Peisach E, Persikova I, Poon BK, Sobolev OV, Ulrich EL, Velankar S, Vonrhein C, Westbrook J, Wojdyr M, Yokochi M, Young JY. Announcing mandatory submission of PDBx/mmCIF format files for crystallographic depositions to the Protein Data Bank (PDB). Acta Crystallogr D Struct Biol. 2019 Apr 1;75(Pt 4):451-454. doi:, 10.1107/S2059798319004522. Epub 2019 Apr 8. PMID:30988261 doi:http://dx.doi.org/10.1107/S2059798319004522