Bacterial Intein-Like Domains (BILs)
From Proteopedia
Bacterial Intein-Like Domains (BILs)
Bacterial intein-like domains (BILs) are auto-catalytic protein domains. BILs are ~140 amino acids long, and have characteristic sequence motifs of the Hint (Hog/ INTein) domain. These domains are highly modular, being integrated in variable positions of non-conserved protein hosts, some of which are extracellular. Similarly to other types of Hint domains, BIL domains may contribute to their host variability through several post-translational biological activities like protein-splicing, self-cleavage and ligation. BILs may also be involved in maturation and conjugation of poly-Ubiqutin-like precursor proteins.
Most BIL domains are present in diverse bacterial species, yet recently they were also found in two ciliates. BILs are divided into two distinct families (A- and B-type) according to their unique sequence motifs. A-type BIL domains catalyze protein-splicing, and of N'- or C'-terminal cleavages, and B-type BIL domains can autocleave at either their N', C' or both ends.
A-type BILs contain similar protein-splicing motifs and active-site residues as inteins. However, these domains can protein-splice even without a C' nucleophilic residue (Cys, Ser or Thr), which is absolutely neccesary for inteins protein-splicing mechanism.
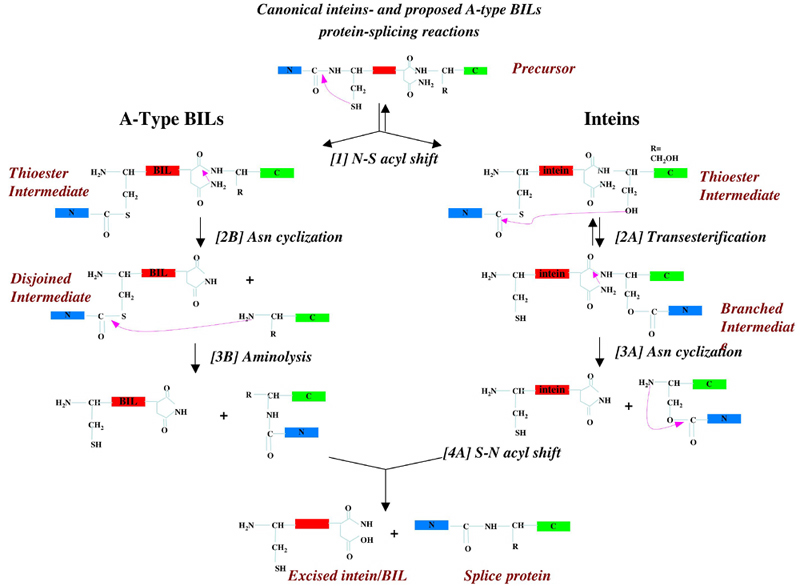 Several BIL domains were experimentally investigated, and showed protein-splicing and self-cleavage activities. An A-type BIL from Pseudomonas syringae (with a +1 Thr) showed protein-splicing and C'-cleavage activities (Amitai et al, 2003). However, an A-type BIL from Magnetospirillum magnetotacticum (with a +1 Tyr) showed N'-cleavage activity, and its protein-splicing activity could be rescued only by reversion of Tyr+1 to Cys (Southworth, 2004). Another A-type BIL from Clostridium thermocellum (with a +1 Ala) showed both protein-splicing, N'- and C'-cleavage activities (Dassa et al, 2004). Two B-type BILs from Rhodobacter sphaeroides showed either N' or C'-cleavage activities, and one B-type BIL from Rhodobacter capsulatus was not found to be active (Dassa et al, 2004).
Several BIL domains were experimentally investigated, and showed protein-splicing and self-cleavage activities. An A-type BIL from Pseudomonas syringae (with a +1 Thr) showed protein-splicing and C'-cleavage activities (Amitai et al, 2003). However, an A-type BIL from Magnetospirillum magnetotacticum (with a +1 Tyr) showed N'-cleavage activity, and its protein-splicing activity could be rescued only by reversion of Tyr+1 to Cys (Southworth, 2004). Another A-type BIL from Clostridium thermocellum (with a +1 Ala) showed both protein-splicing, N'- and C'-cleavage activities (Dassa et al, 2004). Two B-type BILs from Rhodobacter sphaeroides showed either N' or C'-cleavage activities, and one B-type BIL from Rhodobacter capsulatus was not found to be active (Dassa et al, 2004).
The biological function of BILs is yet unknown. BILs, integrated in nonconserved and hypervariable extracellular proteins, can increase its host protein variability by generating alternative products from a single protein precursor: the precursor itself, N- or C-terminal- cleavage products, both N- and C-terminal- flanks, and the splicing product. This can be advantagous in evading the immune system of multi-cellular organisms infected by pathogens, or in adhering the cell to new targets by protein ligation.
"BUBL genes"- BILs in ciliates
BILs were recently found in polyubiquitin-like (Ubl) genes in two ciliates (Tetrahymena thermophila and Paramecium tetraurelia). These genes include tandem repeats of Ubl domains interspersed with BIL domains. This is the only known occurance of more than one BIL domain in the same open-reading frame. Also, this is the only known example of BIL domains in non-bacterial species and in such a unique protein context. Therefore, we propose an autoprocessing role for the BIL domains in maturation and conjugation of the poly-Ubl precursor proteins.
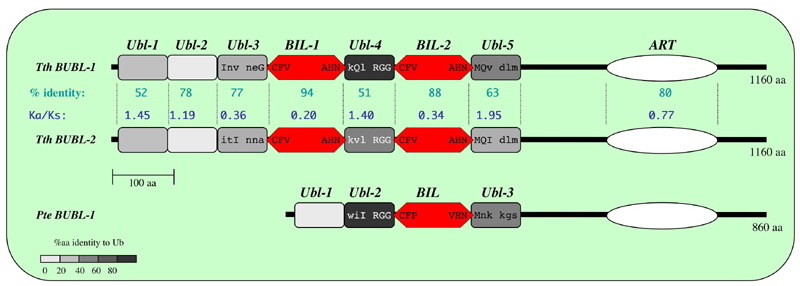 BUBL BIL domains may both cleave their flanking Ubl domains and conjugate them to a new protein or to another moiety. This cis activity might be an alternative to the typical trans maturation and conjugation of Ub and Ubl domains, and could also have enabled a new type of modulation of Ubl function (Dassa et al, 2004).
--Bareket.dassa 15:16, 29 November 2007 (IST)
BUBL BIL domains may both cleave their flanking Ubl domains and conjugate them to a new protein or to another moiety. This cis activity might be an alternative to the typical trans maturation and conjugation of Ub and Ubl domains, and could also have enabled a new type of modulation of Ubl function (Dassa et al, 2004).
--Bareket.dassa 15:16, 29 November 2007 (IST)
